Module: D_GO:0045892
Name = negative regulation of transcription, DNA-dependent
Definition = Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-dependent transcription.
Response module--------
Number of tumors that hit the module: 292
Number of genes in the module: 31
Genes in the module: ADIPOQ;
ATF3;
BMP2;
CAV1;
CD36;
CITED1;
EDN1;
EDNRB;
EGR1;
FABP4;
FGF2;
FOSB;
HOXA2;
HOXA7;
ID4;
KLF4;
LEP;
MEIS2;
NFIB;
NR0B1;
OSR1;
PDE2A;
PPARG;
PROX1;
RASD1;
SFRP1;
SMYD1;
TCEAL7;
TP63;
ZBTB16;
ZFP36;
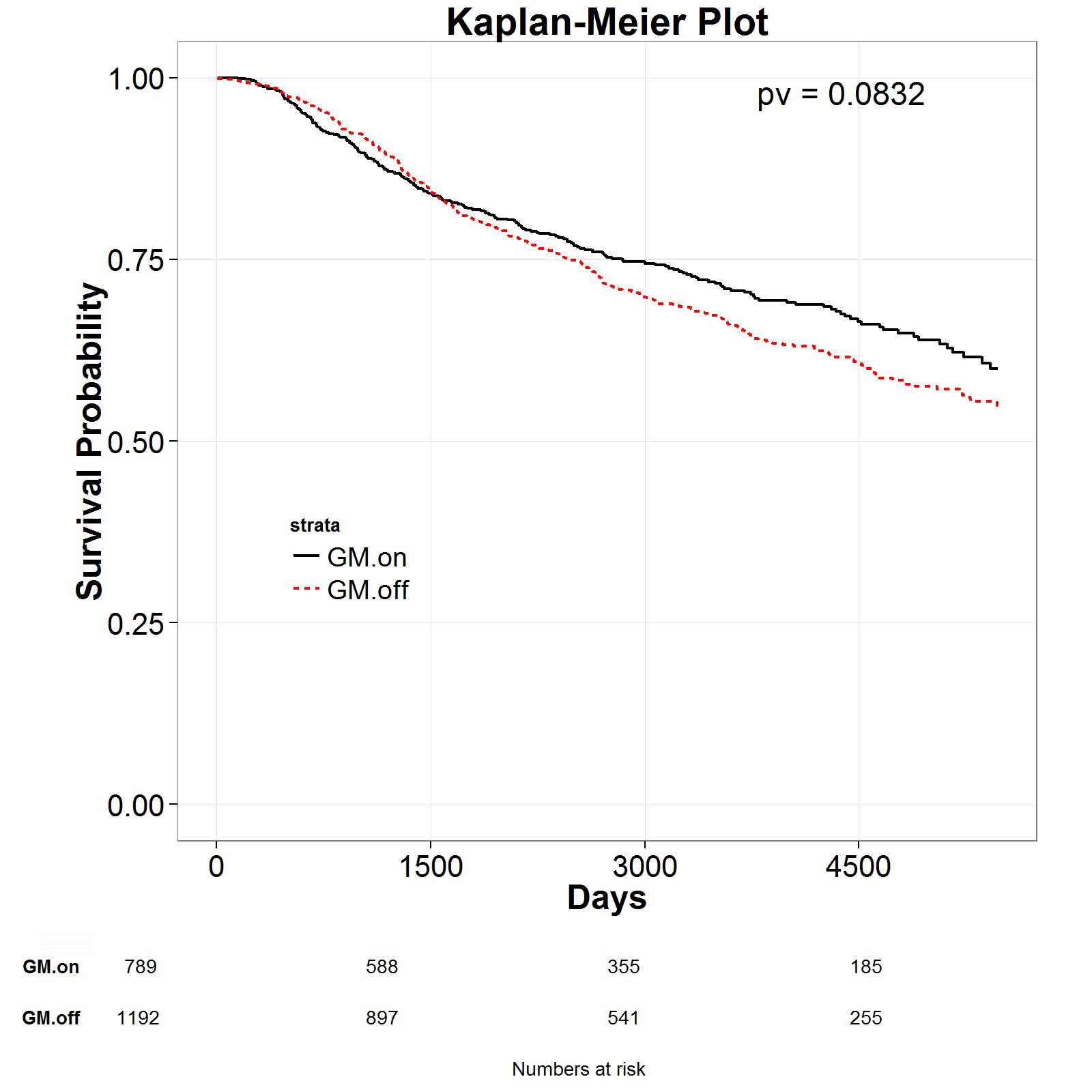
Modu-hit surivval risk:-------
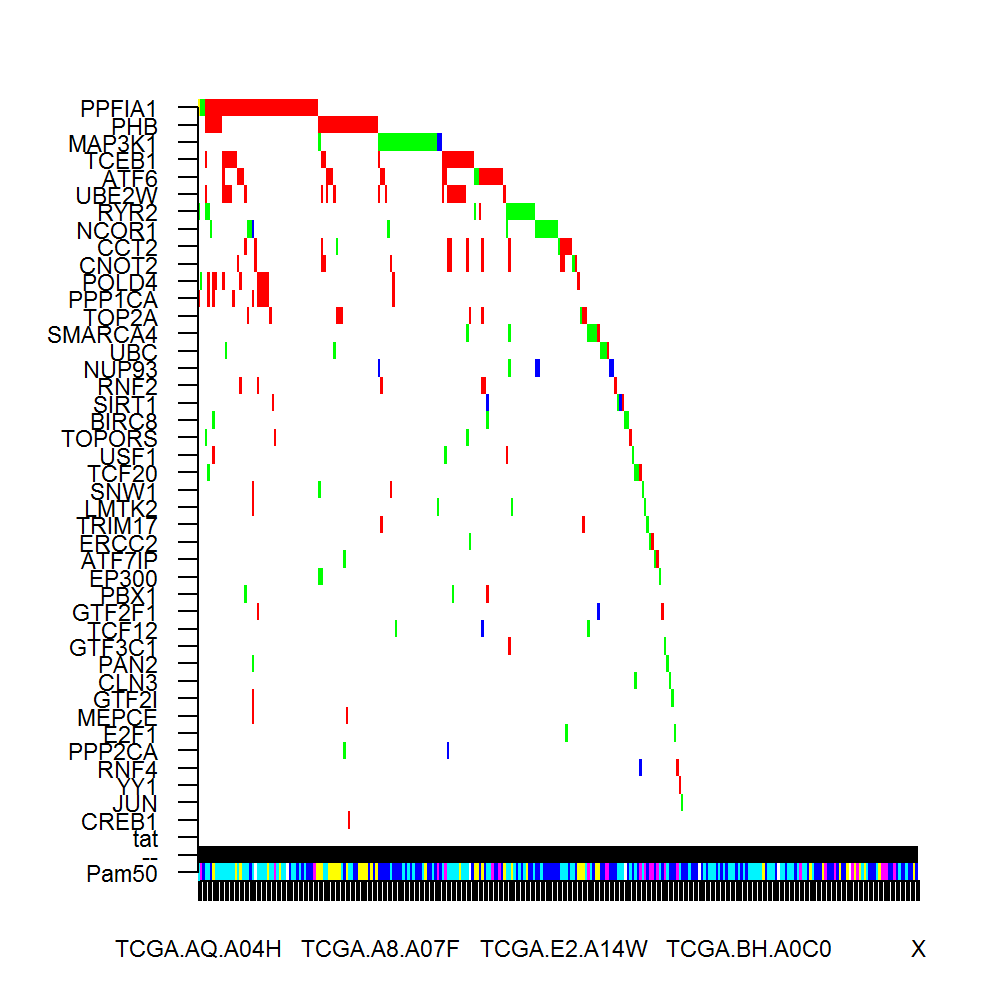
Perturbation module-------
Number of genes in the module: 43
Genes in the module: PPFIA1(3s, 47c);
PHB(31c);
MAP3K1(25s, 2c);
TCEB1(23c);
ATF6(2s, 21c);
UBE2W(21c);
RYR2(16s, 1c);
NCOR1(14s, 1c);
CCT2(2s, 13c);
CNOT2(1s, 13c);
POLD4(1s, 12c);
PPP1CA(11c);
TOP2A(1s, 9c);
SMARCA4(6s, 1c);
UBC(5s, 1c);
NUP93(1s, 5c);
RNF2(6c);
SIRT1(1s, 4c);
BIRC8(4s);
TOPORS(2s, 2c);
USF1(2s, 2c);
TCF20(3s, 1c);
SNW1(2s, 2c);
LMTK2(3s, 1c);
TRIM17(1s, 2c);
ERCC2(2s, 1c);
ATF7IP(2s, 1c);
EP300(3s);
PBX1(2s, 1c);
GTF2F1(3c);
TCF12(2s, 1c);
GTF3C1(1s, 1c);
PAN2(2s);
CLN3(2s);
GTF2I(1s, 1c);
MEPCE(2c);
E2F1(2s);
PPP2CA(1s, 1c);
RNF4(2c);
YY1(1c);
JUN(1s);
CREB1(1c);
tat;
Color Codes: