Module: U_GO:0006281
Name = DNA repair
Definition = The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.
Response module--------
Number of tumors that hit the module: 248
Number of genes in the module: 21
Genes in the module: BLM;
BRIP1;
CDCA5;
CHEK1;
CLSPN;
DTL;
EXO1;
FANCA;
FANCI;
FOXM1;
NEIL3;
POLQ;
PTTG1;
RAD51;
RAD54L;
RECQL4;
TOP2A;
TRIP13;
TYMS;
UBE2T;
UHRF1;
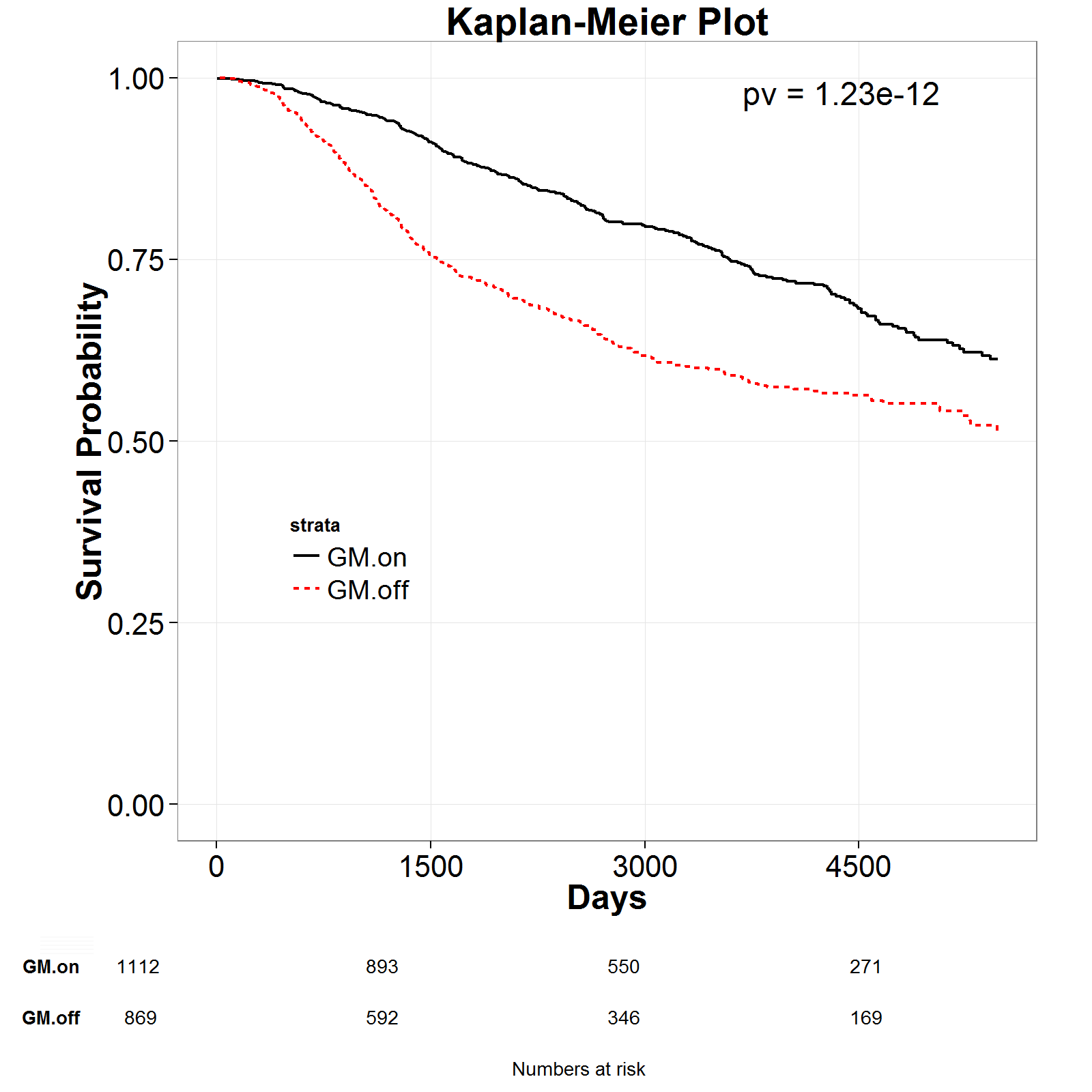
Modu-hit surivval risk:-------
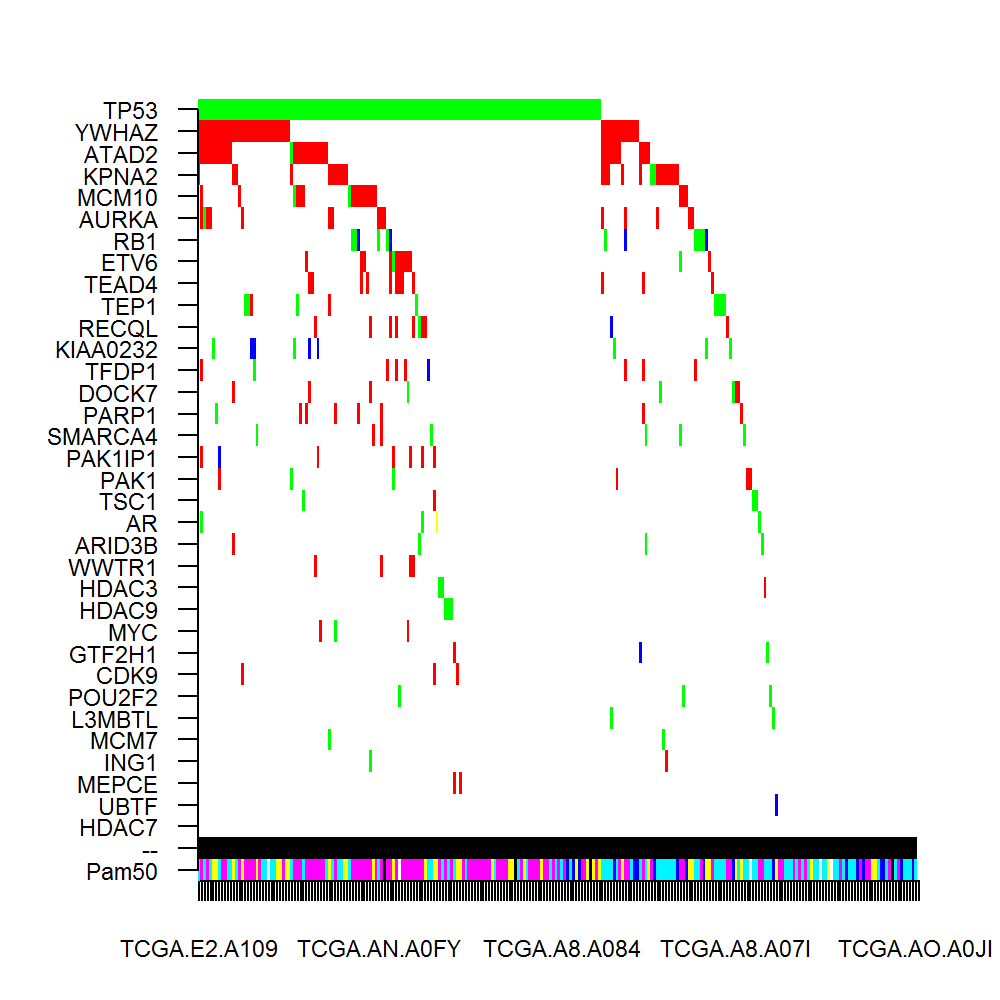
Perturbation module-------
Number of genes in the module: 34
Genes in the module: TP53(139s);
YWHAZ(45c);
ATAD2(1s, 35c);
KPNA2(2s, 24c);
MCM10(2s, 17c);
AURKA(1s, 14c);
RB1(9s, 4c);
ETV6(2s, 11c);
TEAD4(12c);
TEP1(8s, 2c);
RECQL(1s, 9c);
KIAA0232(5s, 4c);
TFDP1(1s, 8c);
DOCK7(3s, 5c);
PARP1(1s, 7c);
SMARCA4(5s, 2c);
PAK1IP1(7c);
PAK1(2s, 4c);
TSC1(3s, 1c);
AR(4s, 1c);
ARID3B(3s, 1c);
WWTR1(4c);
HDAC3(2s, 1c);
HDAC9(3s);
MYC(1s, 2c);
GTF2H1(1s, 2c);
CDK9(3c);
POU2F2(3s);
L3MBTL(2s);
MCM7(2s);
ING1(1s, 1c);
MEPCE(2c);
UBTF(1c);
HDAC7;
Color Codes: